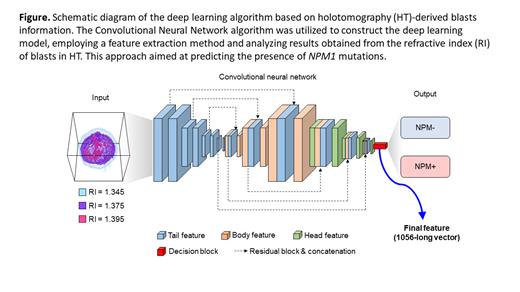
Introduction:
As the clinical significance of genetic mutations becomes clearer, diagnostic classifications are continuously evolving, making judicious decisions about implementing costly genetic testing for diagnosis crucial.
Intriguing observations suggest that Myelodysplastic Syndrome (MDS) or MDS/Myeloproliferative Neoplasm with NPM1 mutation can rapidly progress to Acute Myeloid Leukemia (AML), leading to ambiguity in acute leukemia diagnosis. Consequently, this finding has led to ambiguity surrounding the diagnostic criteria for acute leukemia with low blast counts.
Holotomography (HT) is a cutting-edge microscope that utilizes refractive index (RI) as a contrast medium. It not only provides 2D or 3D images but also offers numerical information about every pixel of cells, promising a more objective diagnosis. Previous studies have successfully distinguished mature cells and blasts using HT. In this study, our aim is to construct a sophisticated deep learning model using data from HT to predict the presence of NPM1 mutations in blasts. This research endeavor holds the potential to enable early detection of these mutations in hematologic malignancies and facilitate the effective utilization of valuable medical resources.
Methods:
We utilized HT to analyze blasts of AML with mutated NPM1 (experimental group) and AML not otherwise specified (control group) but exhibiting similar morphological characteristics from May 2021 to May 2022. We analyzed a total of 648 blasts from 15 patients in the experimental group and 762 blasts from 24 patients in the control group. Cell segmentation for Rule-based features was performed using ilastik version 1.3.3 and MATLAB 2022a. After the initial segmentation using ilastik's machine learning pixel classification feature, further refinements were made in MATLAB, based on prior knowledge of the size and topology of each structure. Subsequently, volumetric and morphological information was measured within these segments.
The cell data of each patient were divided into a training set and a test set. The deep learning algorithm, built using the training set, was then validated with data from the test set. For the deep learning and prediction tasks, we employed a Convolutional Neural Network algorithm (Figure). The classification process involved utilizing both the Rule-based features and the entire feature set, where a linear kernel Support Vector Machine was used. Additionally, we employed the Ensemble average approach, where multiple cells' scores (numerical values associated with the probabilities of each classification) were averaged, and the classification with the highest score was selected as the final prediction.
Finally, the deep learning model was validated using previously unused test set data through the Receiver Operating Characteristic (ROC) curve.
Results:
The dry mass of the nucleolus was significantly increased in AML, NOS; Exp, 0.29341 ± 0.014499 pg; Control, 0.71184 ± 0.033521 pg ( P<0.05). The dry mass of the cytoplasm was higher in AML with mutated NPM1, showing a higher value than that of the nucleolus; Exp, 45.7334 ± 0.50003 pg; Control, 42.9807 ± 0.63882 pg (P<0.05).
When validating the deep learning model constructed based on individual cells, the area under the ROC curve (AUC) was found to be 0.8836. Even within the same individual, the cell morphology can be heterogeneous. Therefore, when the data from 5 cells were treated as a single entity for validating the deep learning model, the AUC was 0.9944.
Conclusions:
It is widely acknowledged that the NPM1 protein, typically localized in the nucleolus, undergoes relocation to the cytoplasm as a result of NPM1 mutation. In this investigation, we devised an advanced deep learning model leveraging numerical data derived from HT, enabling us to prognosticate variations in protein expression based on NPM1 mutations. The HT numerical data exhibited distinctive variations in both nuclear and cytoplasmic analyses contingent on NPM1 mutations.
Given the current uncertainty in leukemia diagnosis, partly attributable to the costly selection of genetic testing, we are optimistic that the integration of HT with our sophisticated deep learning model will pave the way for early diagnosis prediction before conducting additional genetic tests.
Disclosures
No relevant conflicts of interest to declare.